"structRNAfinder"
| chr23_64 | ||
cmsearch-Rfam |
||
Family :tRNA |
Id:RF00005 |
Type:Gene; tRNA; |
Taxonomy:Eukaryota; Bacteria; Viruses; Archaea; | Ontology:GO:0030533 triplet codon-amino acid adaptor activity |
 |
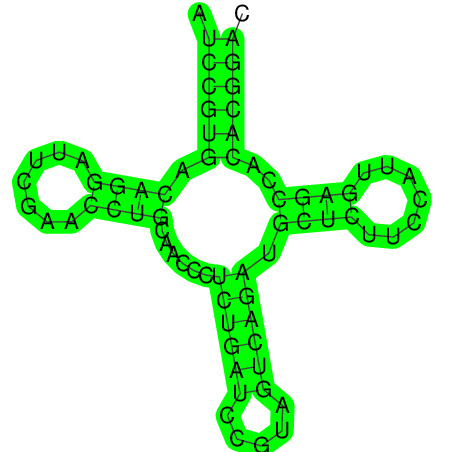
Description:Transfer RNA (tRNA) molecules are approximately 80 nucleotides in length. Their secondary structure includes four shortdouble-helical elements and three loops (D, anti-codon, and T loops). Further hydrogen bonds mediate the characteristic L-shaped molecular structure. tRNAs have two regions of fundamental functional importance: the anti-codon, which is responsible for specific mRNA codon recognition, and the 3' end, to which the tRNAs corresponding amino acid is attached (by aminoacyl-tRNA synthetases). tRNAs cope with the degeneracy of the genetic code in two manners: having more than one tRNA (with a specific anti-codon) for a particular amino acid; and 'wobble' base-pairing, i.e. permitting non-standard base-pairing at the 3rd anti-codon position. | ||
Score:64.7 |
E-value:5.6e-13 |
|
From sequence:215819 |
To sequence:215748 |
|
Alignments: NC | ||
RNAfold |
||
| >chr23_64 AUCCGUGACAGGAUUCGAACCUGCAACCCUCUGAUCCGUAGUCAGAUGCU CUUCCAUUGAGCCACACGGAC .((((((.((((.......))))......((((((.....)))))).((((.......))))..)))))). (-22.50) |
 |
|
Length sequence: 71 |
Length match:70 |
|