"structRNAfinder"
| ssrA | ||
cmscan-Rfam |
||
Family :tmRNA |
Id:RF00023 |
Type:Gene; |
Taxonomy:Eukaryota; Bacteria; Viruses; | Ontology:GO:0006401 RNA catabolic process |
 |
Description:Bacterial tmRNA, also known as 10Sa RNA or SsrA, is named for its dual tRNA andmRNA-like properties. Its role is to liberate the mRNA from a stalled ribosome. To accomplish this part of the tmRNA is used as a reading frame that ends in a translation stop signal. The broken mRNA is replaced in the ribosome by the tmRNA and translation of the tmRNA leads to addition of a proteolysis tag to the incomplete protein enabling recognition by a protease. Recently a number of permuted tmRNA genes have been found encoded in two parts. tmRNAs have been identified in eubacteria and some chloroplasts but are absent from archaeal and eukaryote nuclear genomes. | ||
Score:221.6 |
E-value:9.2e-66 |
|
From sequence:106 |
To sequence:466 |
|
Alignments: v NC | ||
RNAfold |
||
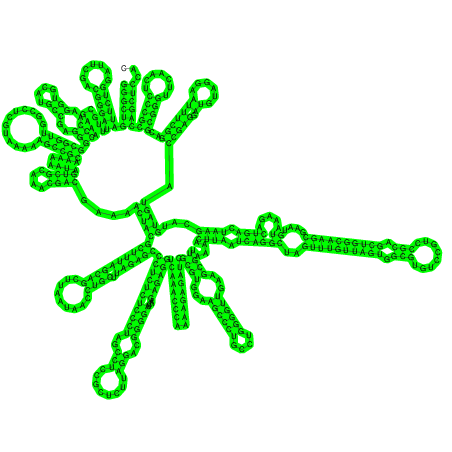
| >ssrA GGGCUGAUUCUGGAUUCGACGGGAUUUGCGAAACCCAAGGUGCAUGCCGA GGGGCGGUUGGCCUCGUAAAAAGCCGCAAAAAAUAGUCGCAAACGACGAA AACUACGCUUUAGCAGCUUAAUAACCUGCUUAGAGCCCUCUCUCCCUAGC CUCCGCUCUUAGGACGGGGAUCAAGAGAGGUCAAACCCAAAAGAGAUCGC GUGGAAGCCCUGCCUGGGGUUGAAGCGUUAAAACUUAAUCAGGCUAGUUU GUUAGUGGCGUGUCCGUCCGCAGCUGGCAAGCGAAUGUAAAGACUGACUA AGCAUGUAGUACCGAGGAUGUAGGAAUUUCGGACGCGGGUUCAACUCCCG CCAGCUCCAC ((((((((((((.......))))))........(((..(((....)))..)))((((((............))))))........((((....))))....(((((((((((((((.........)))).))))))(((((((((((..(((........)))..)))))....))))))..................((((...((((((....))))))...)))).....((((.((((((..((((((((((.(((........))).))))))))))....)).....)))).))))...))))).(((((.((......)))))))..(((((.......)))))))))))... (-116.90) |
 |
|
Length sequence: 360 |
Length match:359 |
|